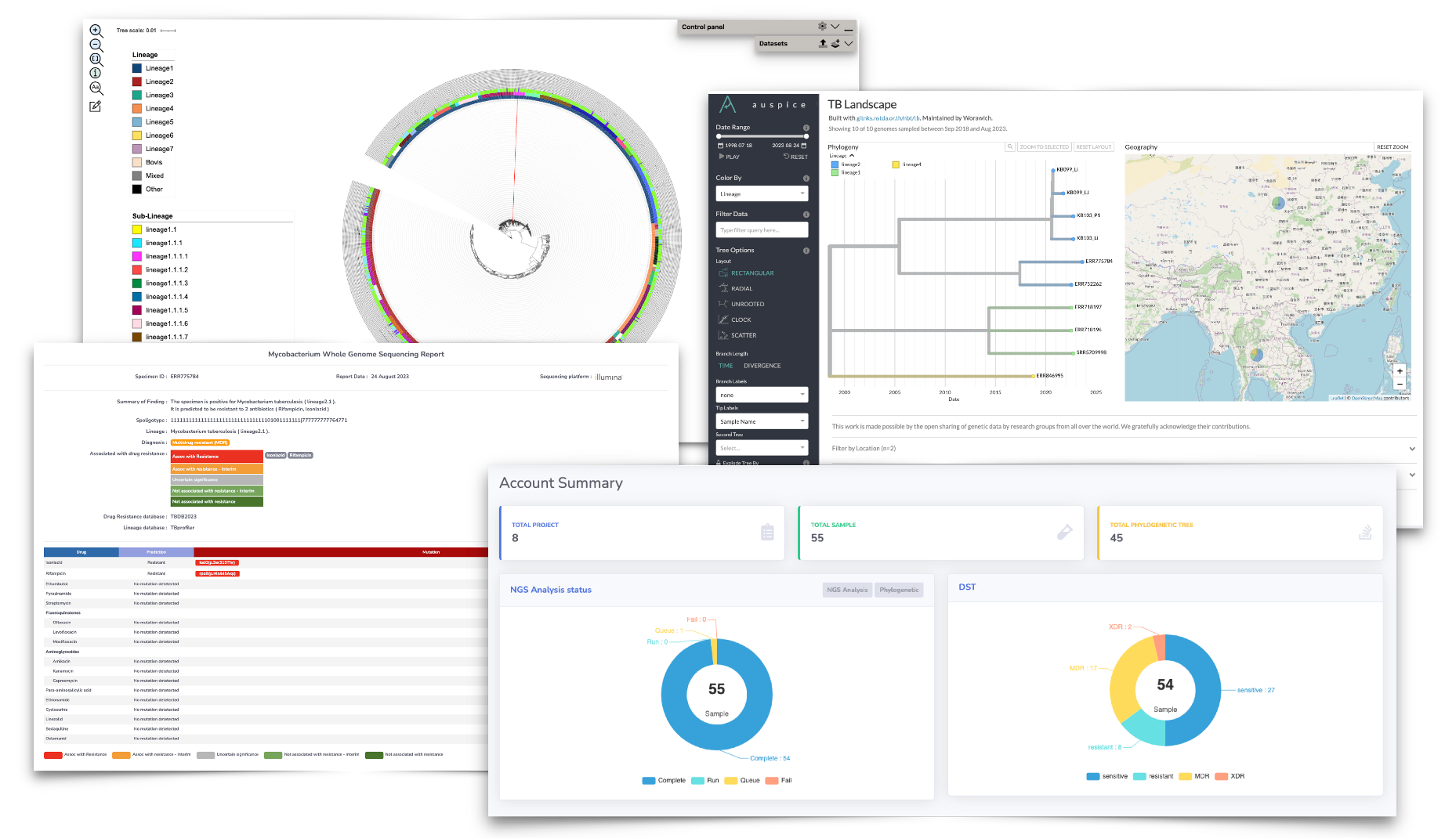
Mycobacterium tuberculosis (MTB) causes a major public health problem. With the high prevalence of MTB in Thailand, the World Health Organization (WHO) has historically identified Thailand as one of the high tuberculosis (TB) burden countries, a group characterized by a significant number of cases across TB infection, co-infection with HIV (TB/HIV), and multidrug-resistant TB (MDR-TB). The increasing incidence of drug-resistant TB significantly hampers the success of TB control programs due to higher treatment failure rates. Often, newly diagnosed TB patients are prescribed multiple drugs empirically because standard TB drug sensitivity testing can take up to two months. Such practices can inadvertently promote a higher incidence of TB drug resistance. Consequently, the WHO recommends the use of TB whole-genome sequencing (WGS) within standard TB control programs, encompassing diagnosis, prediction of TB drug resistance, and TB spread management. WGS offers advantages in terms of speed and cost-effectiveness, making it increasingly a standard practice in public health. However, TB WGS generates large sequencing data files, requiring complex bioinformatic operations that can be challenging for TB interpretation personnel. Addressing this challenge, we are developing a computational platform to assist in accurate TB drug resistance prediction using Next-Generation Sequencing (NGS) analysis. The resulting computational prediction of drug-resistant TB aims to promote the construction of an up-to-date national TB genomic portal, offering a prototype for TB drug programs in Thailand.